Researchers from the Academy of Mathematics and Systems Science (AMSS) of the Chinese Academy of Sciences have teamed up with Stanford University and Tsinghua University scientists to successfully model data on gene regulation with paired expression and chromatin accessibility (PECA) and have developed new tools to infer context-specific regulatory networks.
A gene regulatory network describes the biological process of transcription factor protein binding in the regulatory region of the target gene, which affects its spatial and temporal expression.
Different human tissues and cell types are derived from the same DNA sequence. However, only about half of the genes are transcribed in a particular tissue or cell type, resulting in different morphologies and functionalities for each cell type.
Since the emergence of high-throughput gene expression experiments, computational biologists have been interested in the inference of gene regulatory relationships from gene expression data across diverse cellular contexts corresponding to diverse cell types and experimental conditions.
Interactions among chromatin regulators, sequence specific transcription factors, and cis-regulatory sequence elements are the main driving forces shaping context-specific chromatin structure and gene expression.
However, because of the large number of such interactions, direct data on them are often missing in most cellular contexts.
The present work shows that by modeling matched expression and accessibility data across diverse cellular contexts, it is possible to recover a significant portion of the information in the missing data on binding locations and chromatin states and to achieve accurate inferences for gene regulatory relationships.
The transcriptional regulatory network inferred by PECA provides a detailed view of how trans- and cis-regulatory elements work together to affect gene expression in a context-specific manner.
The researchers proposed a statistical approach based on PECA data across diverse cellular contexts. They highlighted the critical role of chromatin in the regulation of gene expression.
They analyzed the PECA data from the mouse Encyclopedia of DNA Elements (ENCODE) and explored the interpretation of genetic variants relevant to traits and diseases.
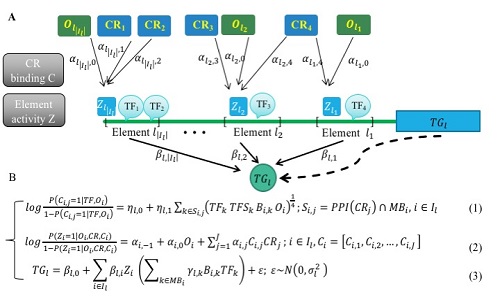
Fig.1. Schematic overview of PECA model (Image by Yong Wang)
Contact:
Yong Wang
Email:ywang@amss.ac.cn
Academy of Mathematics and Systems Science, CAS
http://www.pnas.org/content/early/2017/06/01/1704553114.abstract